Local Lengthscale GP with PyMC
Contents
Local Lengthscale GP with PyMC¶
Author: Zeel B Patel, Nipun Batra
In this chapter, we explore a non-stationary GP discussed by [PKB08] with PyMC.
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import rc
import pymc3 as pm
from sklearn.cluster import KMeans
import warnings
from multiprocessing import Pool
warnings.filterwarnings('ignore')
rc('font', size=16)
Let us define both stationary and non-statioanry RBF kernels,
### Stationary GP
def kernel(a, b, lenghtscale, std):
"""
Borrowed from Nando De Freita's lecture code
https://www.cs.ubc.ca/~nando/540-2013/lectures/gp.py
"""
sqdist = np.square(a - b.T)
return std**2*np.exp(-.5 * (1/lenghtscale) * sqdist)
### LLS GP
def global_kernel(x1, x2, l1, l2, std):
sqdist = np.square(x1 - x2.T)
l1l2meansqr = (np.square(l1)[:, np.newaxis, :] + np.square(l2)[np.newaxis, :, :]).squeeze()/2
# print(sqdist.shape, l1l2meansqr.shape)
return std**2 * pm.math.matrix_dot(np.sqrt(l1),np.sqrt(l2.T)) * (1/np.sqrt(l1l2meansqr)) * np.exp(-sqdist/l1l2meansqr)
def local_kernel(x1, x2, lengthscale):
"""
Borrowed from Nando De Freita's lecture code
https://www.cs.ubc.ca/~nando/540-2013/lectures/gp.py
"""
sqdist = np.square(x1 - x2.T)
return np.exp(-.5 * (1/lengthscale) * sqdist)
We will test the efficacy of these models on a step function data.
n_train = 21
np.random.seed(1234)
# Generate data
def f(X): # target function
return np.sin(5*X) + np.sign(X)
X = np.random.uniform(-1, 1, (n_train, 1)).reshape(-1,1) # data
Y = f(X)[:, 0] + np.random.normal(0,0.2,n_train)
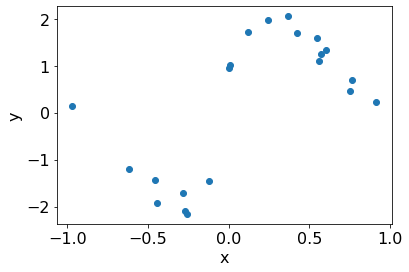
plt.scatter(X, Y);
plt.xlabel('x');
plt.ylabel('y');
Stationary GP model¶
basic_model = pm.Model()
with basic_model:
# Priors for unknown model parameters
# Variance
kernel_std = pm.Lognormal("kernel_std", 0, 0.1)
# Length scale
kernel_ls = pm.Lognormal("kernel_ls", 0, 1)
noise_sigma = pm.Lognormal("noise_sigma", 0, 1)
K = kernel(X, X, kernel_ls, kernel_std)
K += np.eye(X.shape[0]) * np.power(noise_sigma, 2)
y = pm.MvNormal("y", mu = 0, cov = K, observed = Y.squeeze())
pm.model_to_graphviz(basic_model.model)
Let us find MAP estimate of paramaters.
map_estimate = pm.find_MAP(model=basic_model)
map_estimate
{'kernel_std_log__': array(0.02655438),
'kernel_ls_log__': array(-2.68569403),
'noise_sigma_log__': array(-1.27853804),
'kernel_std': array(1.02691009),
'kernel_ls': array(0.06817386),
'noise_sigma': array(0.27844408)}
Now, we will sample from the posterior.
with basic_model:
# draw 2000 posterior samples per chain
trace = pm.sample(2000,return_inferencedata=False,tune=2000)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [noise_sigma, kernel_ls, kernel_std]
Sampling 4 chains for 2_000 tune and 2_000 draw iterations (8_000 + 8_000 draws total) took 15 seconds.
import arviz as az
az.plot_trace(trace);
Now, we will infer the \(y\) values at new input locations.
test_x = np.linspace(-1.5, 1.5, 100).reshape(-1, 1)
train_x = X
train_y = Y
def post(train_x, train_y, test_x, kernel, kernel_ls, kernel_std, noise):
N = len(train_x)
K = kernel(train_x, train_x, kernel_ls, kernel_std)+noise**2*np.eye(len(train_x))
N_star = len(test_x)
K_star = kernel(train_x, test_x, kernel_ls, kernel_std)
K_star_star = kernel(test_x, test_x, kernel_ls, kernel_std)
posterior_mu = K_star.T@np.linalg.inv(K)@(train_y)
posterior_sigma = K_star_star - K_star.T@np.linalg.inv(K)@K_star
# Instead of size = 1, we can also sample multiple times given a single length scale, kernel_std and noise
return np.random.multivariate_normal(posterior_mu, posterior_sigma, size=1)
# Make predictions at new locations.
train_y = Y.squeeze()
n_samples = 500
preds = np.stack([post(train_x, train_y, test_x=test_x, kernel=kernel, kernel_ls=trace['kernel_ls'][b],
kernel_std=trace['kernel_std'][b],
noise=trace['noise_sigma'][b])
for b in range(n_samples)])
ci = 95
ci_lower = (100 - ci) / 2
ci_upper = (100 + ci) / 2
preds_mean = preds.reshape(n_samples, len(test_x)).mean(0)
preds_lower = np.percentile(preds, ci_lower, axis=0)
preds_upper = np.percentile(preds, ci_upper, axis=0)
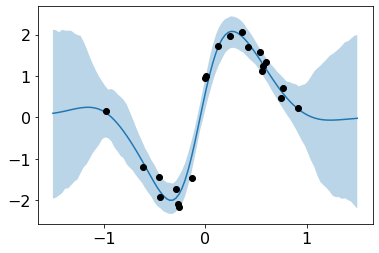
plt.plot(test_x,preds.reshape(n_samples, len(test_x)).mean(axis=0))
plt.scatter(train_x, train_y, c='black', zorder=3, label='data')
plt.fill_between(test_x.flatten(), preds_upper.flatten(), preds_lower.flatten(), alpha=.3, label='95% CI');
LLS GP¶
We will learn local lengthscales at 3 input locations choosen by KMeans clustering.
n_local = 3
lls_model = pm.Model()
param_X = KMeans(n_local, random_state=0).fit(X).cluster_centers_
with lls_model:
### Local GP
# local lengthscale
local_ls = pm.Lognormal("local_ls", 0, 1)
param_ls = pm.Lognormal("param_ls", 0, 1, shape=(n_local, 1))
local_K = local_kernel(param_X, param_X, local_ls)
local_K_star = local_kernel(X, param_X, local_ls)
### global GP
# global lengthscales
global_ls = pm.math.exp(pm.math.matrix_dot(local_K_star, pm.math.matrix_inverse(local_K), pm.math.log(param_ls)))
# global variance
global_std = pm.Lognormal("global_std", 0, 1)
# global noise
global_noise_sigma = pm.Lognormal("global_noise_sigma", 0, 1)
global_K = global_kernel(X, X, global_ls, global_ls, global_std)
global_K += np.eye(X.shape[0])*global_noise_sigma**2
y = pm.MvNormal("y", mu = 0, cov = global_K, observed = Y)
pm.model_to_graphviz(lls_model.model)
Let us find MAP estimate of paramaters.
NSmap_estimate = pm.find_MAP(model=lls_model)
NSmap_estimate
{'local_ls_log__': array(-0.88024402),
'param_ls_log__': array([[-0.28197464],
[-0.05336731],
[-1.54950142]]),
'global_std_log__': array(0.09507128),
'global_noise_sigma_log__': array(-1.61986434),
'local_ls': array(0.41468171),
'param_ls': array([[0.75429281],
[0.94803173],
[0.21235382]]),
'global_std': array(1.09973724),
'global_noise_sigma': array(0.19792555)}
with lls_model:
# draw 2000 posterior samples per chain
trace = pm.sample(4000,return_inferencedata=False,tune=2000)
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [global_noise_sigma, global_std, param_ls, local_ls]
Sampling 4 chains for 2_000 tune and 4_000 draw iterations (8_000 + 16_000 draws total) took 43 seconds.
There were 12 divergences after tuning. Increase `target_accept` or reparameterize.
There were 13 divergences after tuning. Increase `target_accept` or reparameterize.
There were 18 divergences after tuning. Increase `target_accept` or reparameterize.
There were 14 divergences after tuning. Increase `target_accept` or reparameterize.
import arviz as az
with lls_model:
az.plot_trace(trace);
Let us predict the values at new locations.
test_x = np.linspace(-1.5, 1.5, 100).reshape(-1, 1)
train_x = X
train_y = Y
def post(local_ls, param_ls, global_std, global_noise):
N = len(train_x)
param_K_inv = np.linalg.inv(local_kernel(param_X, param_X, local_ls))
local_K = local_kernel(train_x, param_X, local_ls)
global_ls = np.exp(local_K@param_K_inv@param_ls)
local_K_star = local_kernel(test_x, param_X, local_ls)
global_ls_star = np.exp(local_K_star@param_K_inv@param_ls)
K = global_kernel(train_x, train_x, global_ls, global_ls, global_std)+np.eye(N)*global_noise**2
K_inv = pm.math.matrix_inverse(K)
K_star = global_kernel(train_x, test_x, global_ls, global_ls_star, global_std)
posterior_mu = pm.math.matrix_dot(K_star.T,K_inv,train_y)
return posterior_mu.eval()
# Instead of size = 1, we can also sample multiple times given a single length scale, kernel_std and noise
return np.random.multivariate_normal(posterior_mu.eval(), posterior_sigma.eval(), size=1)
# Make predictions at new locations.
n_samples = 100
preds = np.stack([post(local_ls=trace['local_ls'][b],
param_ls=trace['param_ls'][b],
global_std=trace['global_std'][b],
global_noise=trace['global_noise_sigma'][b])
for b in range(n_samples)])
Let us visualize predictive mean and variance.
ci = 95
ci_lower = (100 - ci) / 2
ci_upper = (100 + ci) / 2
preds_mean = preds.reshape(n_samples, len(test_x)).mean(0)
preds_lower = np.percentile(preds, ci_lower, axis=0)
preds_upper = np.percentile(preds, ci_upper, axis=0)
plt.plot(test_x,preds.reshape(n_samples, len(test_x)).mean(axis=0))
plt.scatter(train_x, train_y, c='black', zorder=3, label='data')
plt.fill_between(test_x.flatten(), preds_upper.flatten(), preds_lower.flatten(), alpha=.3, label='95% CI');
for x_loc in param_X:
plt.vlines(x_loc, -3, 3);
plt.vlines(x_loc, -3, 3, label='Latent locations');
plt.legend(bbox_to_anchor=(1,1));
plt.ylim(-2.5,2.5);
plt.xlabel('x')
plt.ylabel('y');
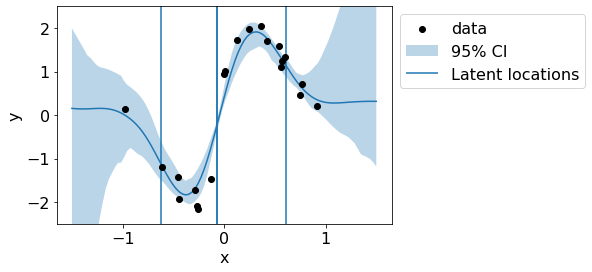
Let us visualize the latent lengthscales.
local_ls = NSmap_estimate['local_ls']
param_ls = NSmap_estimate['param_ls']
local_K = local_kernel(param_X, param_X, local_ls)
local_K_star = local_kernel(test_x, param_X, local_ls)
global_ls = pm.math.exp(pm.math.matrix_dot(local_K_star, pm.math.matrix_inverse(local_K), pm.math.log(param_ls)))
plt.plot(test_x, global_ls.eval(), label='Local lengthscales', color='r');
plt.vlines(param_X.squeeze(), *plt.ylim(), label='latent locations')
plt.xlabel('x')
plt.ylabel('lengthscales');
plt.legend(bbox_to_anchor=(1,1));
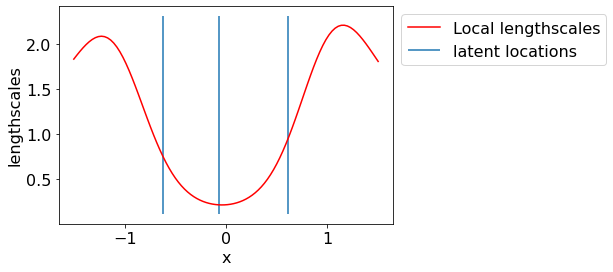
We see that lengthscales are low in the center region where the underlying function is having a sudden jump. Thus, LLS model is giving us sensible predictions.
